In order to develop the Clinical Trial Risk Tool, we had to conduct a quality control exercise on the components. Each parameter which is fed into the complexity model is trained and evaluated independently. For an overview of how the tool works, please read this blog post.
I used two datasets to train and evaluate the tool:
By combining the two datasets I was able to get some of the advantages of a large dataset and some of the advantages of a smaller, more accurate dataset.
For validation on the manual dataset, I used cross-validation. For validation on the ClinicalTrials.gov dataset, I took the third digit of the trial’s NCT ID. Trials with values 0-7 were used for training, with value 8 were used for validation, and those with value 9 are held out as a future test set.
Validation scores on small manually labelled dataset (about 100 protocols labelled, but 300 labelled for number of subjects). You can reproduce my experiments using the notebooks from this folder.
| Component | Accuracy – manual validation dataset | AUC – manual validation dataset | Technique |
| Condition (Naive Bayes: HIV vs TB) | 88% | 100% | Naive Bayes |
| SAP (Naive Bayes) | 85% | 87% | Naive Bayes |
| Effect Estimate | 73% | 95% | Naive Bayes |
| Number of Subjects | 69% (71% within 10% margin) | N/A | Rule based combined with Random Forest |
| Simulation | 94% | 98% | Naive Bayes |
You can reproduce my experiments using the notebooks from this folder. As a sanity check I also trained a Naive Bayes classifier for some of these components to check that our models are outperforming a reasonable baseline.
| Component | Accuracy – ClinicalTrials.gov validation dataset | Baseline Accuracy (Naive Bayes) – ClinicalTrials.gov validation dataset | Technique |
| Phase | 75% | 45% | Ensemble – rule based + random forest |
| SAP | 82% | Naive Bayes | |
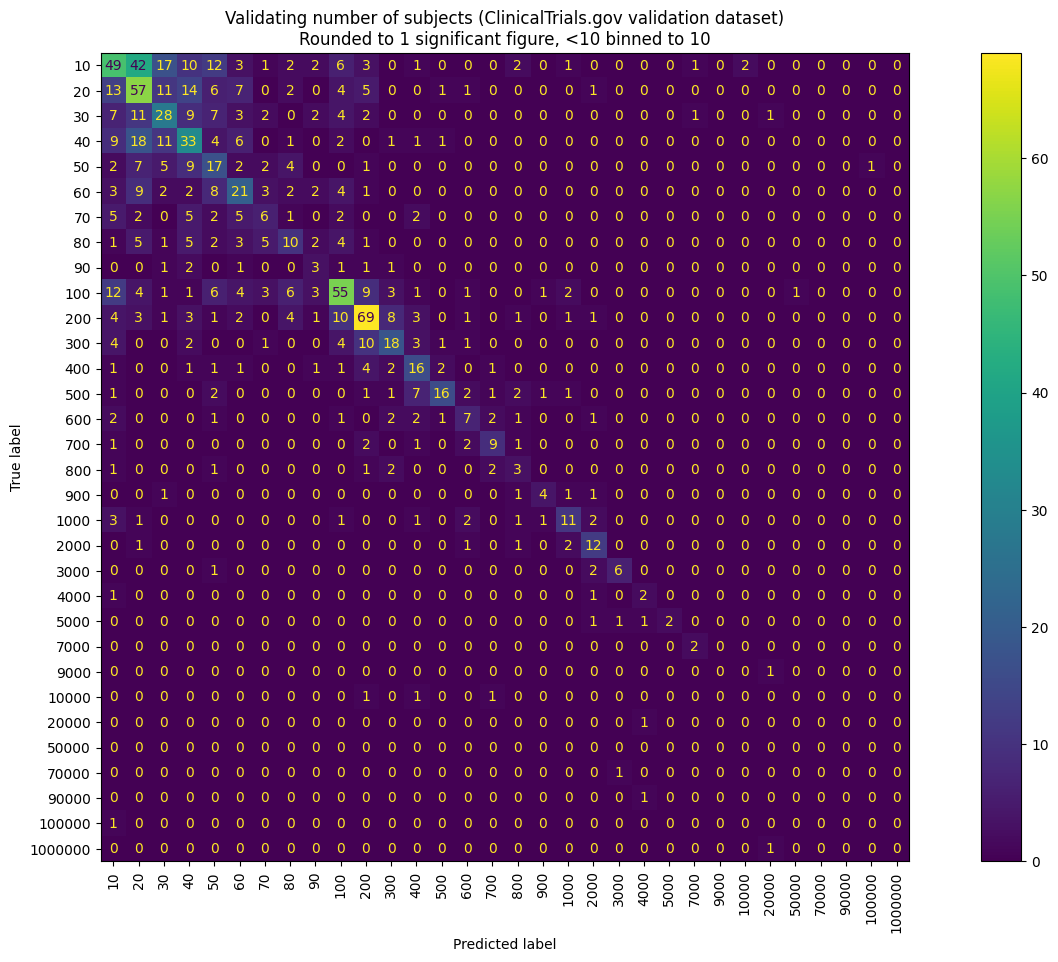
| Number of Subjects | 13% | 6% | Rule based combined with Random Forest |
| Number of Arms | 58% | 52% | Ensemble |
| Countries of Investigation | AUC 87% | N/A | Ensemble – rule based + random forest + Naive Bayes |
In particular I found that the ClinicalTrials.gov value for the sample size was particularly inaccurate, hence the very low performance of the model on that value.
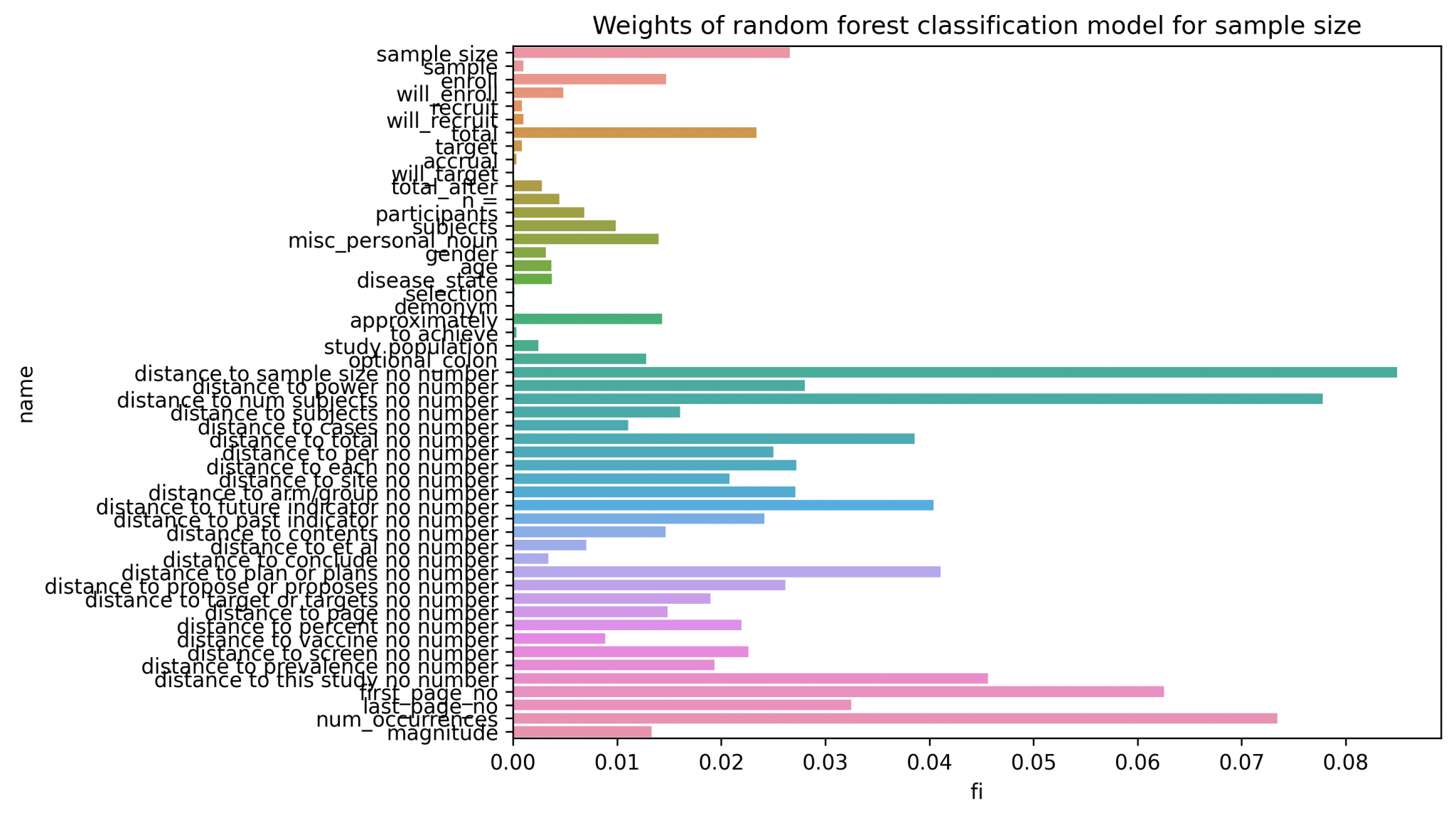
By far the most difficult model was the number of subjects (sample size) classifier.
I designed this component as a stage of manually defined features to identify candidate sample sizes (numeric values in the text), combined with a random forest using these features to identify the most likely candidate. Here is an output of the feature importances of the random forest model.
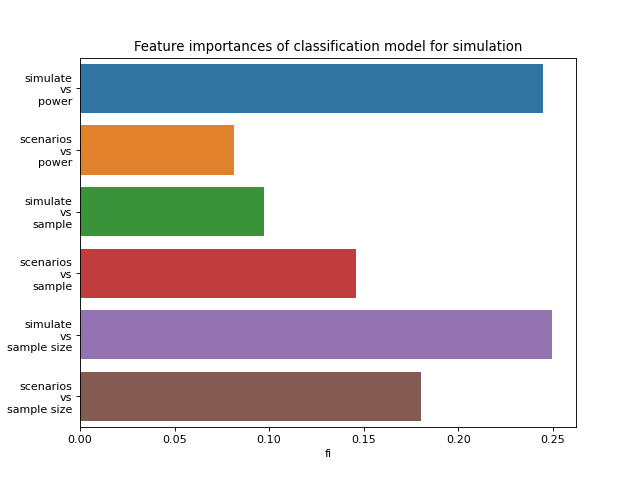
Similarly, the simulation classifier is a random forest that uses manually defined features of key words:
For any of the components, we also plotted a Confusion Matrix.
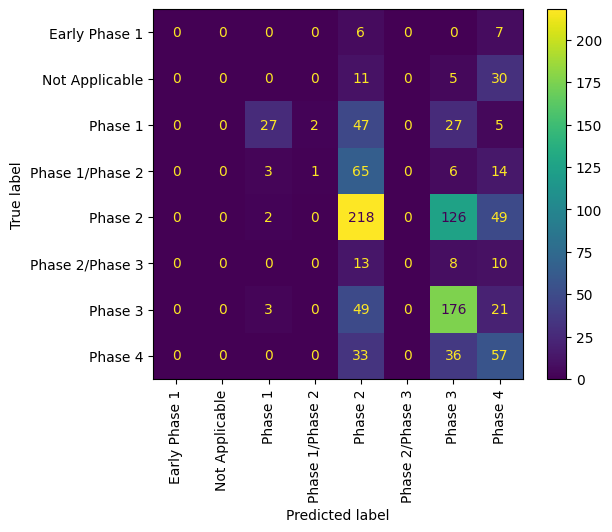
Confusion matrix for the baseline (Naive Bayes) phase extractor on the ClinicalTrials.gov validation dataset
Since each component is designed differently, it has been complex to validate the performance for clinical trial risk assessment.
However I have provided some Jupyter notebooks in the repository to run the validation and reproduce my results.
There is still much scope for improvement of several features, especially sample size.
Some parameters, such as simulation, were not available in the ClinicalTrials.gov dataset and so could only be trained and validated manually. We hope to be able to annotate more data for these areas.

Estimating the total cost of a clinical trial before it runs is challenging. Public data on past trial costs can be hard to come by, as many companies guard this information carefully. Trials in high income countries and low and middle income countries have very different costs. Upload your clinical trial protocol and create a cost benchmark with AI Protocol to cost benchmark The Clinical Trial Risk Tool uses AI and Natural Language Processing (NLP) to estimate the cost of a trial using the information contained in the clinical trial protocol.

You can download a white paper about clinical trial cost benchmarking here Estimating the total cost of a clinical trial before it runs is challenging. Public data on past trial costs can be hard to come by, as many companies guard this information carefully. Trials in high income countries and low and middle income countries have very different costs. Clinical trial costs are not normally distributed.[1] I took a dataset of just over 10,000 US-funded trials.

Guest post by Safeer Khan, Lecturer at Department of Pharmaceutical Sciences, Government College University, Lahore, Pakistan Introduction The success of clinical studies relies heavily on proper financial planning and budgeting. These processes directly impact key factors such as project timelines, resource allocation, and compliance with regulatory requirements. The accurate forecasting of costs for clinical trials, however, is a highly complex and resource-intensive process. A study by the Tufts Center for the Study of Drug Development found that the average cost of developing a new drug is approximately $2.