People have asked us often, how was the Clinical Trial Risk Tool trained? Does it just throw documents into ChatGPT? Or conversely, is it just an expert system, where we have painstakingly crafted keyword matching rules to look for important snippets of information in unstructured documents?
Most of the tool is built using machine learning techniques. We either hand-annotated training data, or took training data from public sources.
The different models inside the Clinical Trial Risk tool have been trained on real data, mostly taken from clinical trial repositories such as clinicaltrials.gov (the US trial database which is maintained by the National Library of Medicine). We have sometimes used data which we have tagged ourselves and sometimes used data points provided by investigators as they uploaded their protocols to the repository.
Where possible, we have used simple approaches such as rule based methods (expert systems) or small language models such as Naive Bayes. For example, classifying a protocol into a disease area is a task which can relatively easily be completed using a Naive Bayes text classifier. Very rarely have we resorted to generative AI (in fact, all generative AI is turned off by default until you actively enable it) and sometimes we have used more complex models or we have used ensemble models which combine more than one machine learning approach or a rule based approach together with a machine learning approach to refine it.
We have kept track of the number of documents used to train each model, so that training can be reproduced. The first iteration of the Clinical Trial Risk Tool has been published in Gates Open Research and so more precise documentation of how how each model is trained is also featured in our publication [1].
Where applicable, we have validated models on an independent test data set or we have used cross-validation, which means that we have used the majority of our data set to train a model and then withheld a portion to test it and then switched the testing portion around and then eventually trained a new model on the entire data set. We report metrics such as accuracy but we primarily work with Area Under the ROC curve (AUC). You can see a breakdown of the individual models, accuracies and AUCs below.
The table below shows the performance of individual models measured in terms of AUC and accuracy, as well as the size of the test set. Where applicable we have usually reported Precision, Recall and F1 score and this data can be provided on request. Most, but not all, of the models and associated training data is open sourced in our Github repository: training scripts and training data.
| Component | Model type | Train docs | Test docs | AUC | Accuracy |
|---|---|---|---|---|---|
| Condition | Naive Bayes multi-label classifier | 1025 | 1025 (cross validation) | 99% | 88% |
| SAP | Ensemble Naive Bayes classifiers | 33 (678 pages) | cross-validation | 87% | 85% |
| Effect estimate | Ensemble: Rule based + Naive Bayes | 58 | 15 | 95% | 73% |
| Sample size | Ensemble: rule based + Random Forest | 282 | cross-validation | N/A | 69% (71% within 10% margin) |
| Countries of investigation | Ensemble rule based + CNN + Random Forest + Naïve Bayes | 9540 | 2385 | 87% | N/A |
| Simulation | Random Forest with feature engineering | 49 | cross-validation | 98% | 94% |
| Number of arms | Ensemble: Naive Bayes plus Convolutional Neural Network | 9538 | 1085 | N/A | 58% |
| Phase | Ensemble: Rule based plus Convolutional Neural Network | 9538 | 1085 | N/A | 75% |
| Drugs | Ensemble: Rule based plus Naive Bayes | 10,000 | 20% of the mentions | 95% | 97% |
| Inclusion and exclusion criteria and endpoints | Named Entity Recognition | 45 (197 pages containing at least one entity) | 2 (17 pages) | N/A | N/A |
| Vaccine | Naive Bayes binary classifier | 145 | 15 | 96% | 87% |
| Child subjects | Naive Bayes binary classifier | 7500 | 2500 | 87% | 87% |
| Radiation | Naive Bayes binary classifier | 975 | 326 | 90% | 95% |
| Trial title | Named Entity Recognition | 18541 (1st page only for each document) | 20 (1st page only for each document) | N/A | N/A |
| Intervention text | Named Entity Recognition | 1603 (1st 15 pages only for each document) | 20 (1st 15 pages only for each document) | N/A | N/A |
| Document type | Naive Bayes multi-label classifier | 14040 (1st 2 pages only for each document) | 3459 (1st 2 pages only for each document) | 98% | 99% |
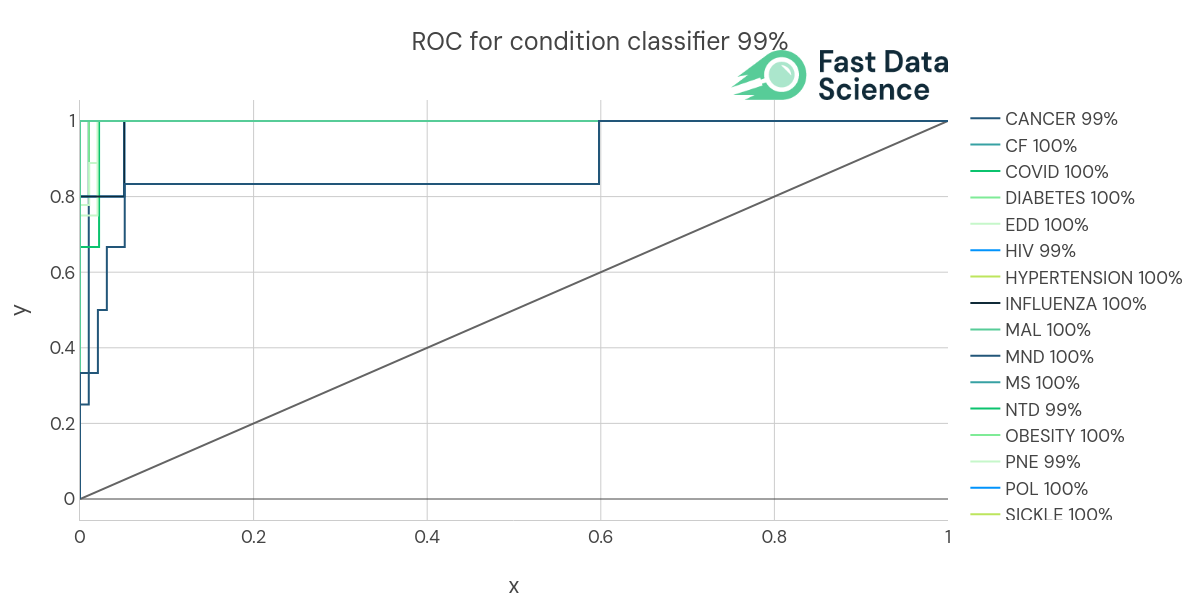
We identify the condition or disease using a Naive Bayes multi-label text classifier. This was trained on 1025 protocols from ClinicalTrials.gov which belonged to the following categories.
| CANCER, CF, COVID, DIABETES, EDD, HIV, HYPERTENSION, INFLUENZA, MAL, MND, MS, NTD, OBESITY, PNE, POL, SICKLE, STROKE, TB, other
We used n-fold cross validation, so we trained a machine learning model on all the documents bar one, and then validated its accuracy on the remaining document. We achieved an AUC (area under the curve) of 99% and an accuracy of 88%.

ROC (Receiver Operating Curve) of the Condition classifier

Confusion matrix of the Condition classifier
We identify whether or not the protocol contains a complete Statistical Analysis Plan using a Naïve Bayes classifier operating on the text of the whole document on word level. In addition, candidate pages which are likely to be part of the SAP are highlighted to the user using a Naïve Bayes classifier operating on the text of each page individually. This model was trained on 33 documents with 578 pages individually annotated.
This model achieved 85% accuracy and 87% AUC.
More information is available in our publication: https://gatesopenresearch.org/articles/7-56/v1 [1].
The training code for the SAP classifier is in our open source repository on Github: https://github.com/fastdatascience/clinical_trial_risk/blob/main/train/train_sap_classifier.py
We identify the presence or absence of an estimated effect size by using a set of regular expressions and patterns to pick out candidate effect sizes, and then a multinomial Naive Bayes classifier to prioritise them. It was trained on 58 hand annotated documents, where the effect estimate (typically a percentage) was identified and tagged by a human, and it achieved 73% accuracy and 95% AUC on validation data consisting of 15 documents.
A rule-based component written in spaCy identifies candidate values for the effect estimate from the numeric substrings present in the document. These can be presented as percentages, fractions, or take other surface forms. A weighted Naive Bayes classifier which is applied to a window of 20 tokens around each candidate number found in the document, and the highest ranking effect estimate candidates are returned. The values are displayed to the user, but only the binary value of the presence or absence of an effect estimate enters into the risk calculation. [1]
The training code for the effect estimate classifier is in our open source repository on Github: https://github.com/fastdatascience/clinical_trial_risk/blob/main/train/train_effect_estimate_classifier.py
We hand-tagged 282 documents with the sample size and wrote a set of manual rules to find candidate sample sizes (typically numbers). We trained a rule-based component written in spaCy to identify candidate values for the sample size from the numeric substrings present in the document. These values are then passed to a random forest classifier, which ranks them by likelihood of being the true sample size, and identifies any substrings such as “per arm” or “per cohort”, which can then be used to multiply by the number of arms if applicable. The model reached 69% accuracy at identifying the number of subjects exactly and 71% accuracy at finding it within a 10% margin. [1]
The training code for the sample size classifier is in our open source repository on Github: https://github.com/fastdatascience/clinical_trial_risk/blob/main/train/train_num_subjects_classifier.py
If you enable generative AI in the tool, we use the OpenAI API to refine the prediction made by our machine learning classifier, in order to make the sample size identification more accurate.
We took 9540 protocols from ClinicalTrials.gov and used these as our main benchmark to train an ensemble of machine learning models to identify the country or countries of investigation. The model combined rule based components to pick out explicitly mentioned country names, and some machine learning, to identify which countries mentioned are really countries of investigation (and not just parts of citations or other false positives). The model achieved AUC 87% on a held out test dataset of 2385 protocols. [1]
We also relied on the Country named entity recognition Python library, developed by Fast Data Science [2].
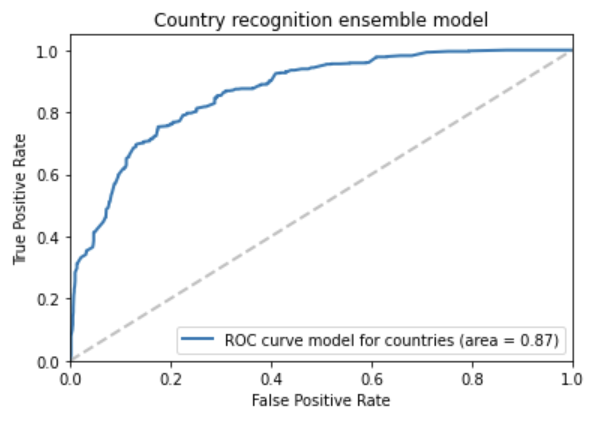
Above: ROC curve of the country ensemble model
The training code for the separate sample size classifiers is in our open source repository on Github:
The Clinical Trial Risk Tool identifies whether simulation was used for sample size determination. This is a Random Forest operating on engineered features extracted from the document. This was trained on 49 documents and evaluated on the same 49 using 49-fold cross-validation (so each document was excluded from the training set individually, the model was retrained, and then it was used to validate the model). The model achieved 94% accuracy and 98% AUC when validated using cross-validation.
Above: Feature importances for the simulation model
The training code for the simulation classifier is in our open source repository on Github:
The number of arms is identified using an AI model, specifically an ensemble machine learning and rule-based tool using the NLP library spaCy and scikit-learn Random Forest. If the number of arms cannot be identified exactly, the neural network attempts to get it approximately right by assigning the number of arms into a bin.
The models were trained on 9,538 protocols from ClinicalTrials.gov and achieved 58% accuracy evaluated on a separate 1085 protocols.
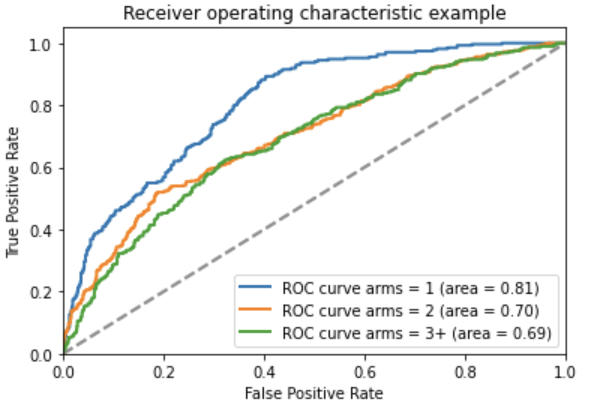
Above: ROC curve of the arms neural network model
The training code for the separate “number of arms” classifiers is in our open source repository on Github:
The trial phase is extracted from the text using an ensemble between a convolutional neural network text classifier, implemented using the NLP library spaCy, and a rule-based pattern matching algorithm combined with a rule-based feature extraction stage and a random forest binary classifier, implemented using Scikit-Learn. Both models in the ensemble output an array of probabilities, which were averaged to produce a final array. The phase candidate returned by the ensemble model was the maximum likelihood value.
Both were trained on trained on 9,538 protocols from ClinicalTrials.gov and evaluated on a separate 1085 protocols. The ensemble model achieved 75% accuracy when evaluated on a separate 1,085 protocols.
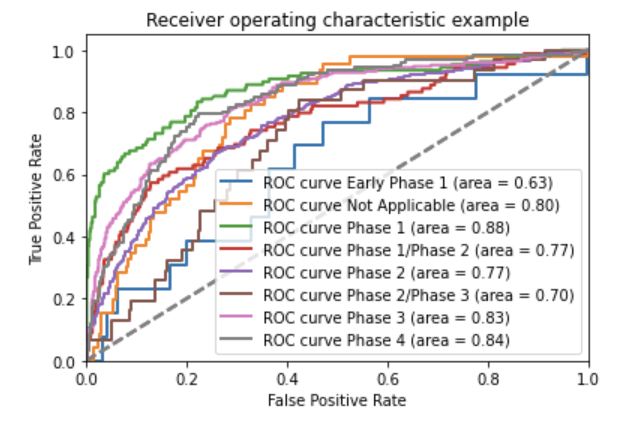
Above: ROC curve of the phase convolutional neural network model

The training code for the separate phase classifiers is in our open source repository on Github:
The drug names are identified using the Drug Named Entity Recognition Python library [3], and then a machine learning model is used to identify whether a particular drug is under investigation or not.
The machine learning model (AI) was trained on 10,000 protocols from ClinicalTrials.gov, which resulted in 262,018 drug mentions. The model and achieved 97% accuracy at identifying the drugs under investigation when evaluated on 79,056 separate drug name mentions.
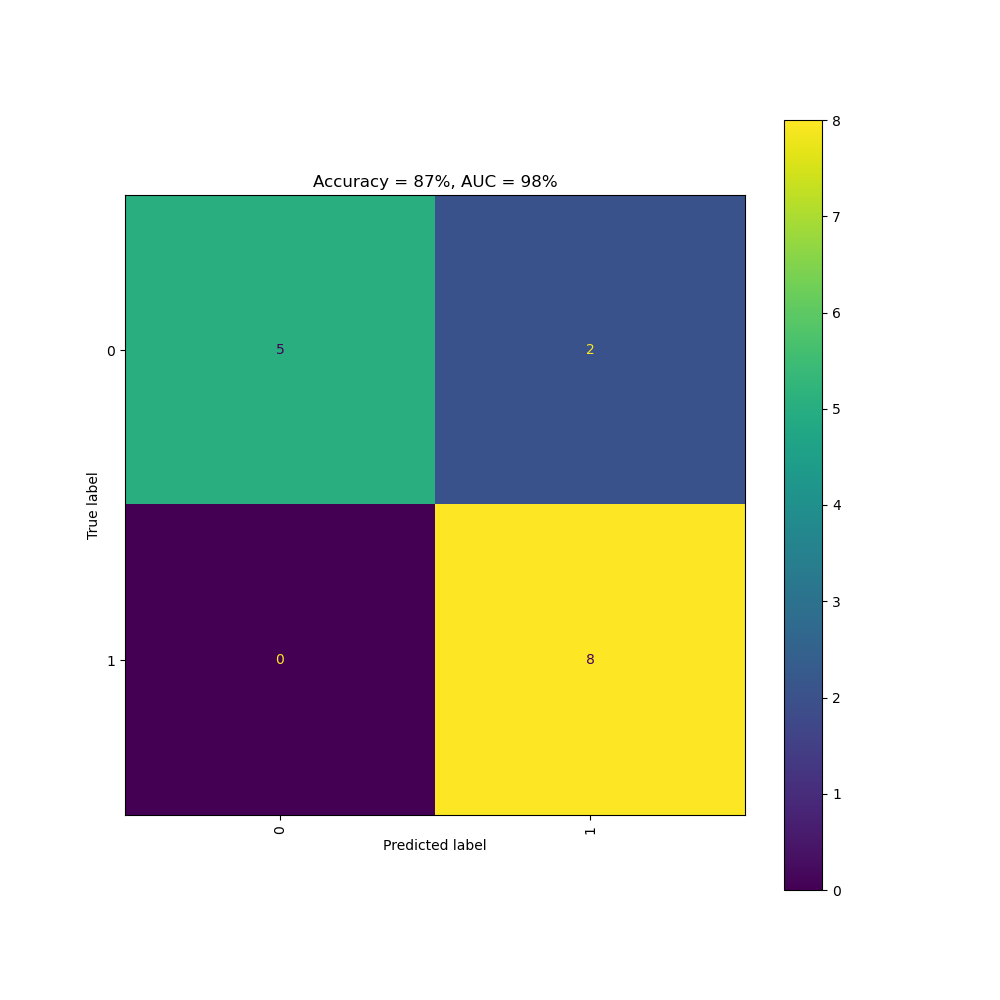
The Clinical Trial Risk Tool determines the likelihood of the trial being a vaccine trial using a Naive Bayes model which was trained on 145 protocols from ClinicalTrials.gov and validated on 15 protocols. The AI achieved 87% accuracy and 86% AUC.
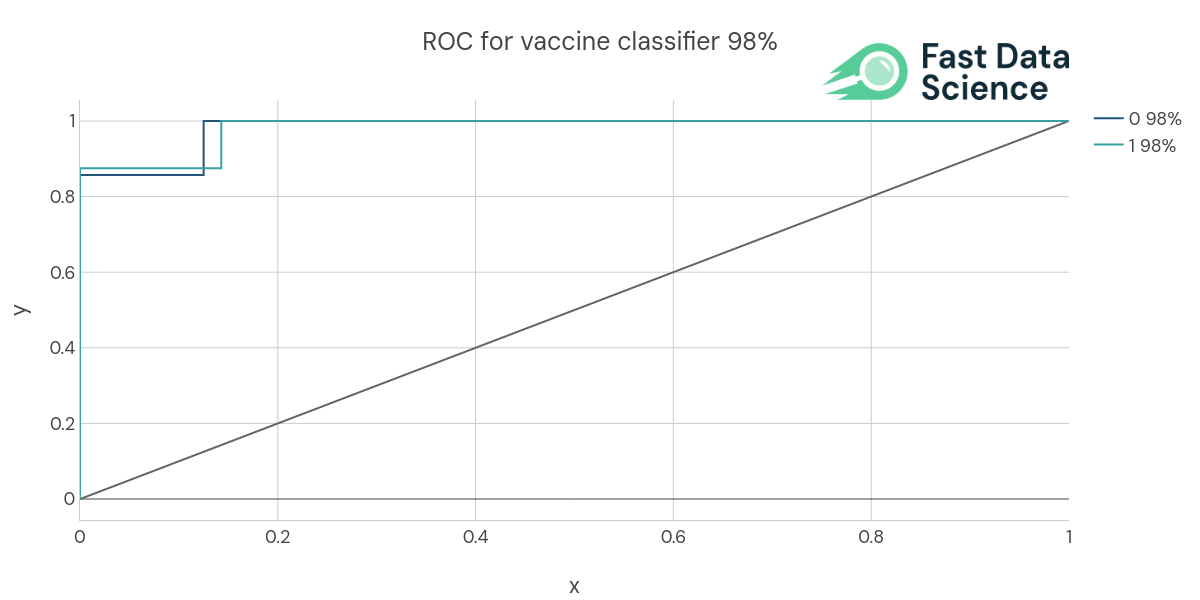
Above: ROC curve of the vaccine model
The Clinical Trial Risk Tool determines the likelihood of the trial involving child subjects using a Naive Bayes model which was trained on 7500 protocols from ClinicalTrials.gov and validated on 2500 protocols. The AI achieved 87% accuracy and 87% AUC.
We use a spaCy named entity recognition model to pick out the inclusion criteria, exclusion criteria, and endpoints from the protocol text. This was trained on 45 hand-tagged documents and validated on 2 documents. We achieved the following score metrics at identifying these three types of entity:
E # LOSS TOK2VEC LOSS NER ENTS_F ENTS_P ENTS_R SCORE
--- ------ ------------ -------- ------ ------ ------ ------
27 5400 54005.78 1530.25 59.87 64.38 55.95 0.60
We use a spaCy named entity recognition model to pick out the trial title. We used data from the ClinicalTrials.gov repository to annotate 18,541 training documents, taking page 1 for each one (assuming the title occurs on the first page). We validated the model on 20 test documents, also taken from ClinicalTrials.gov. We achieved the following score metrics at identifying this type of entity:
E # LOSS TOK2VEC LOSS NER ENTS_F ENTS_P ENTS_R SCORE
--- ------ ------------ -------- ------ ------ ------ ------
0 5000 586.11 488.24 85.00 85.00 85.00 0.85
We use a spaCy named entity recognition model to pick out the trial title. We used data from the ClinicalTrials.gov repository to annotate 1,603 training documents, taking pages 1 to 15 for each one (assuming the title occurs on the first page). We validated the model on 20 test documents, also taken from ClinicalTrials.gov. This resulted in 6290 training pages and 87 test pages. We achieved the following score metrics at identifying this type of entity:
E # LOSS TOK2VEC LOSS NER ENTS_F ENTS_P ENTS_R SCORE
--- ------ ------------ -------- ------ ------ ------ ------
0 6000 2634.65 822.64 42.69 77.72 29.42 0.43
We identify the document type using a Naive Bayes multi-label text classifier. This was trained on 17499 documents (SAPs, ICFs, and protocols) from ClinicalTrials.gov.
We trained the machine learning model on 14040 documents and validated its accuracy on the remaining 3459 documents. We achieved an AUC (area under the curve) of 98% and an accuracy of 91.1%. Document types outside the above set (e.g. synopses) are identified using rules, since it was harder to obtain the same quantity of training documents.
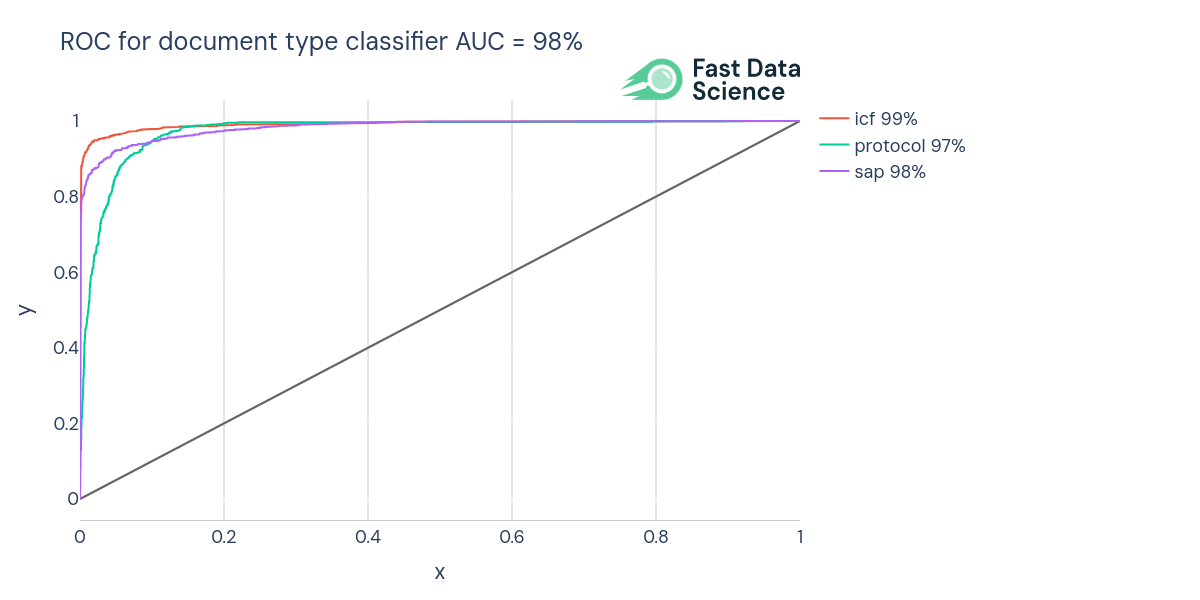
ROC (Receiver Operating Curve) of the document type classifier
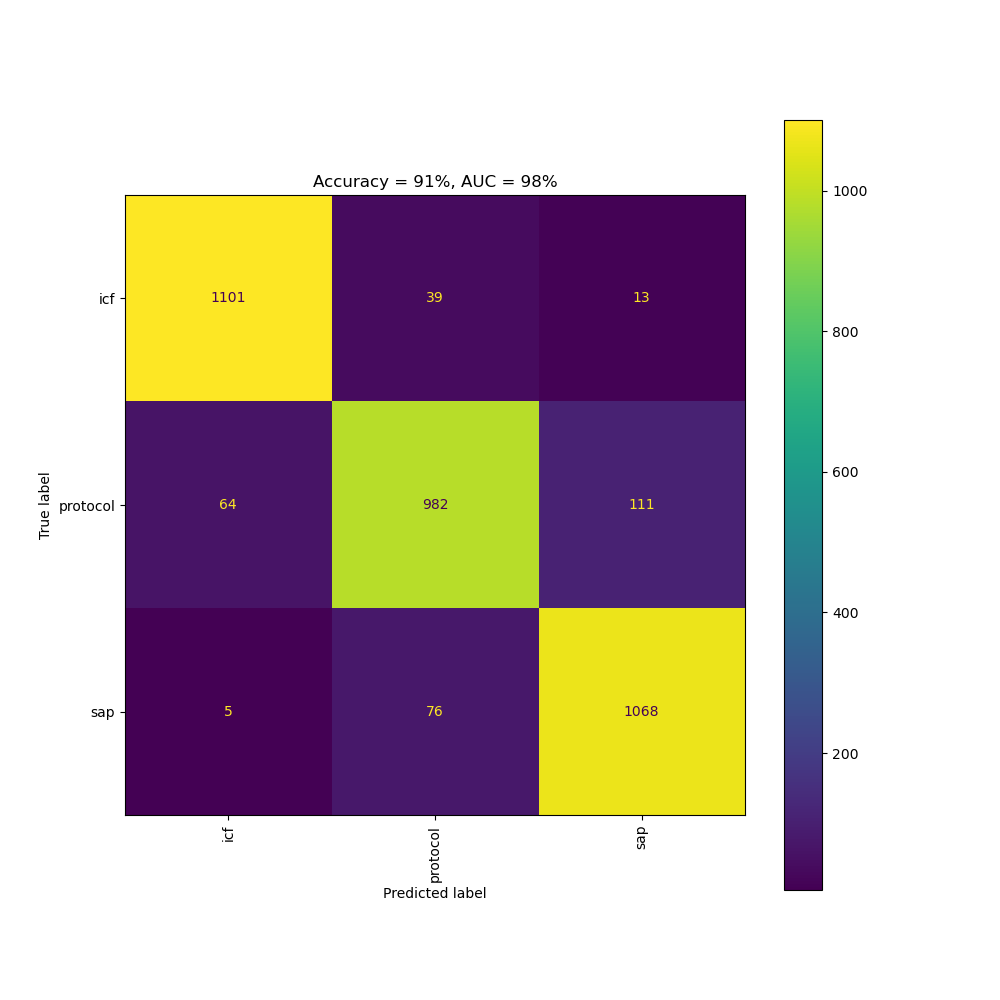
Confusion matrix of the document type classifier
Wood TA and McNair D. Clinical Trial Risk Tool: software application using natural language processing to identify the risk of trial uninformativeness [version 1; peer review: 1 approved with reservations]. Gates Open Res 2023, 7:56 https://doi.org/10.12688/gatesopenres.14416.1
Wood, T.A., Country Named Entity Recognition [Computer software], Version 0.4, accessed at https://fastdatascience.com/country-named-entity-recognition/, Fast Data Science Ltd (2022)
Wood, T.A., Drug Named Entity Recognition [Computer software], Version 1.0.3, accessed at https://fastdatascience.com/drug-named-entity-recognition-python-library, Fast Data Science Ltd (2024)

Estimating the total cost of a clinical trial before it runs is challenging. Public data on past trial costs can be hard to come by, as many companies guard this information carefully. Trials in high income countries and low and middle income countries have very different costs. Upload your clinical trial protocol and create a cost benchmark with AI Protocol to cost benchmark The Clinical Trial Risk Tool uses AI and Natural Language Processing (NLP) to estimate the cost of a trial using the information contained in the clinical trial protocol.

You can download a white paper about clinical trial cost benchmarking here Estimating the total cost of a clinical trial before it runs is challenging. Public data on past trial costs can be hard to come by, as many companies guard this information carefully. Trials in high income countries and low and middle income countries have very different costs. Clinical trial costs are not normally distributed.[1] I took a dataset of just over 10,000 US-funded trials.

Guest post by Safeer Khan, Lecturer at Department of Pharmaceutical Sciences, Government College University, Lahore, Pakistan Introduction The success of clinical studies relies heavily on proper financial planning and budgeting. These processes directly impact key factors such as project timelines, resource allocation, and compliance with regulatory requirements. The accurate forecasting of costs for clinical trials, however, is a highly complex and resource-intensive process. A study by the Tufts Center for the Study of Drug Development found that the average cost of developing a new drug is approximately $2.